<h1>Introduction to tidyverse/R</h1>
<h2>Brandon LeBeau</h2>
<h3>University of Iowa</h3>
# About Me
- I'm an Assistant Professor in the College of Education
+ I enjoy model building, particularly longitudinal models, and statistical programming.
- I've used R for over 10 years.
+ I have 4 R packages, 3 on CRAN, 1 on GitHub
* simglm
* pdfsearch
* highlightHTML
* SPSStoR
- GitHub Repository for this workshop: <https://github.com/lebebr01/iowa_data_science>
# Why teach the tidyverse
- The tidyverse is a series of packages developed by Hadley Wickham and his team at RStudio. <https://www.tidyverse.org/>
- I teach/use the tidyverse for 3 major reasons:
+ Simple functions that do one thing well
+ Consistent implementations across functions within tidyverse (i.e. common APIs)
+ Provides a framework for data manipulation
# PSQF 6250
- Small plug for my online course
- <https://myui.uiowa.edu/my-ui/courses/details.page?_ticket=X-wrjriCUraqNq7N5T9reX3rnNZc97wT&id=848077&ci=148839>
# Course Setup
```r
install.packages("tidyverse")
```
```r
library(tidyverse)
```
```
## -- Attaching packages --------------------------------------- tidyverse 1.2.1 --
```
```
## v ggplot2 2.2.1 v purrr 0.2.4
## v tibble 1.3.4 v dplyr 0.7.4
## v tidyr 0.7.2 v stringr 1.2.0
## v readr 1.1.1 v forcats 0.2.0
```
```
## -- Conflicts ------------------------------------------ tidyverse_conflicts() --
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()
```
# Explore Data
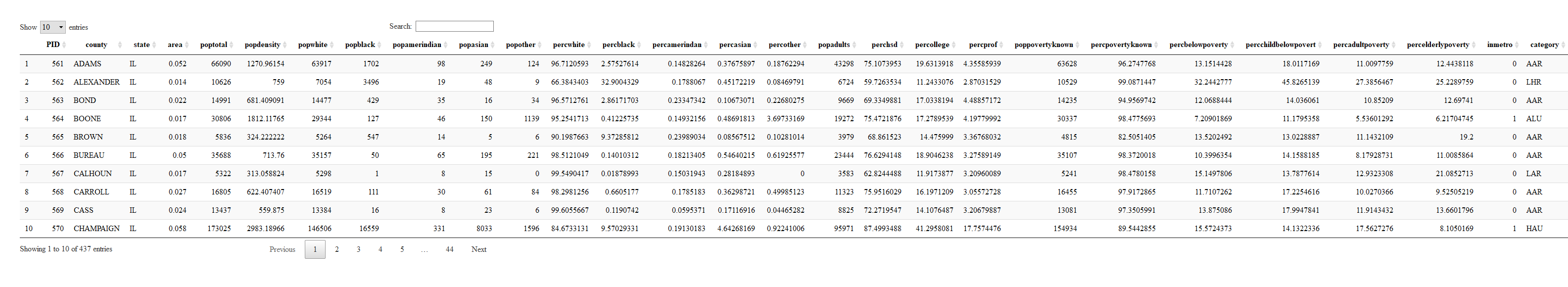
# First ggplot
```r
ggplot(data = midwest) +
geom_point(mapping = aes(x = popdensity, y = percollege))
```
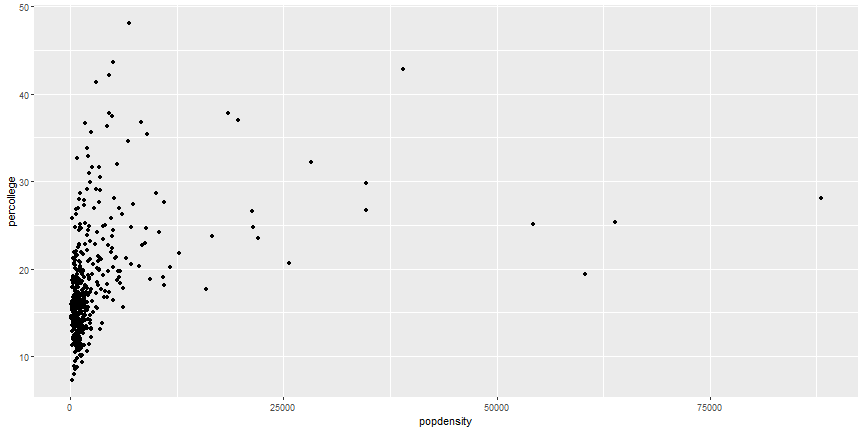
# Equivalent Code
```r
ggplot(midwest) +
geom_point(aes(x = popdensity, y = percollege))
```
# Your Turn
1. Try plotting `popdensity` by `state`.
2. Try plotting `county` by `state`.
+ Does this plot work?
3. Bonus: Try just using the `ggplot(data = midwest)` from above.
+ What do you get?
+ Does this make sense?
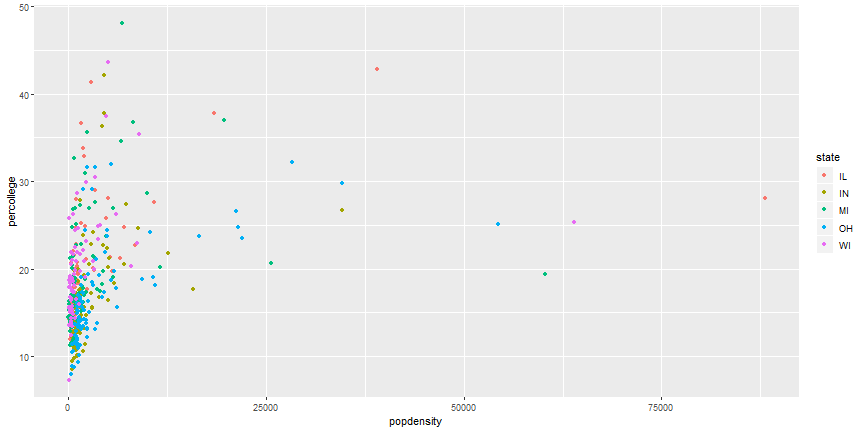
# Add Aesthetics
```r
ggplot(midwest) +
geom_point(aes(x = popdensity, y = percollege, color = state))
```
# Global Aesthetics
```r
ggplot(midwest) +
geom_point(aes(x = popdensity, y = percollege), color = 'pink')
```
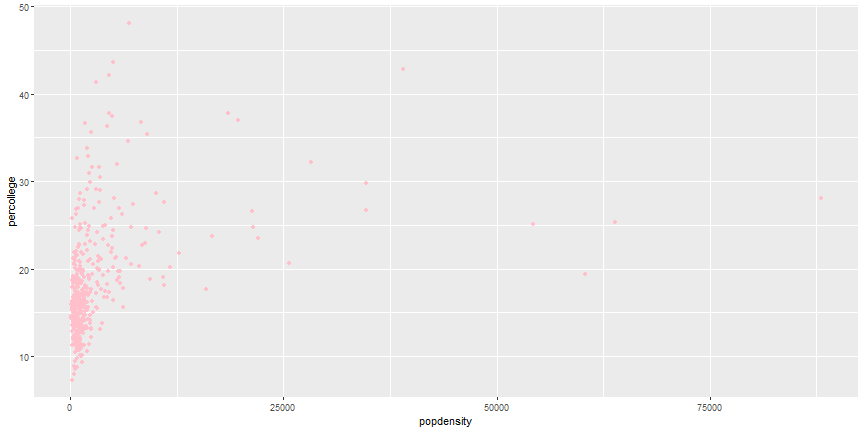
# Your Turn
1. Instead of using colors, make the shape of the points different for each state.
2. Instead of color, use `alpha` instead.
+ What does this do to the plot?
3. Try the following command: `colors()`.
+ Try a few colors to find your favorite.
4. What happens if you use the following code:
```r
ggplot(midwest) +
geom_point(aes(x = popdensity, y = percollege, color = 'green'))
```
# Additional Geoms
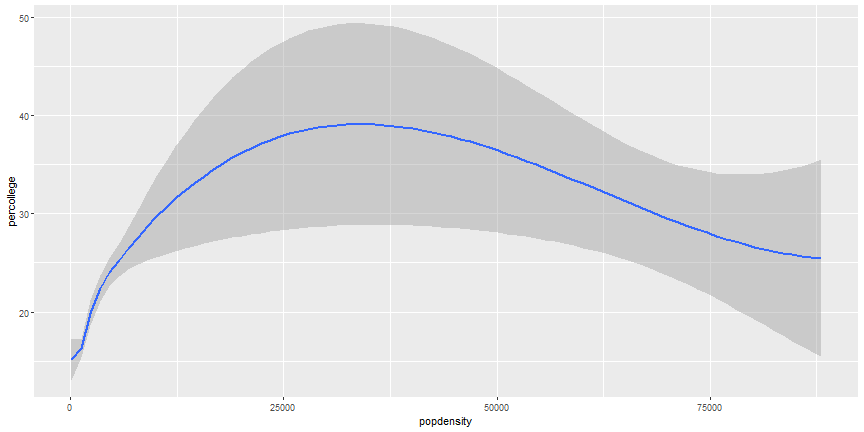
```r
ggplot(midwest) +
geom_smooth(aes(x = popdensity, y = percollege))
```
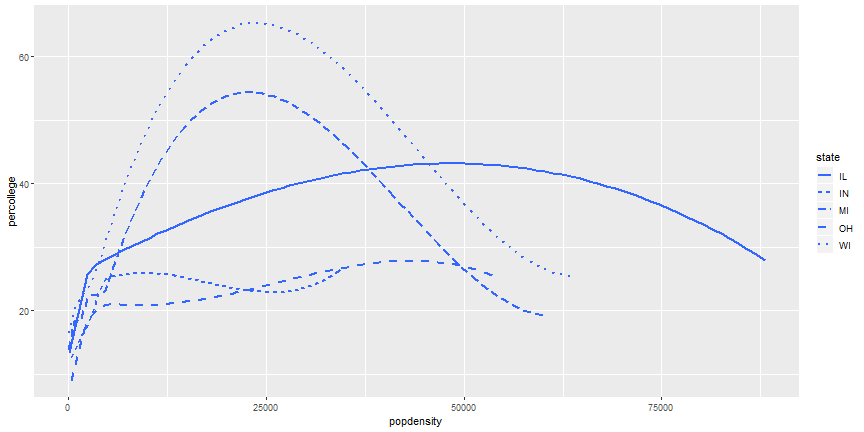
# Add more Aesthetics
```r
ggplot(midwest) +
geom_smooth(aes(x = popdensity, y = percollege, linetype = state),
se = FALSE)
```
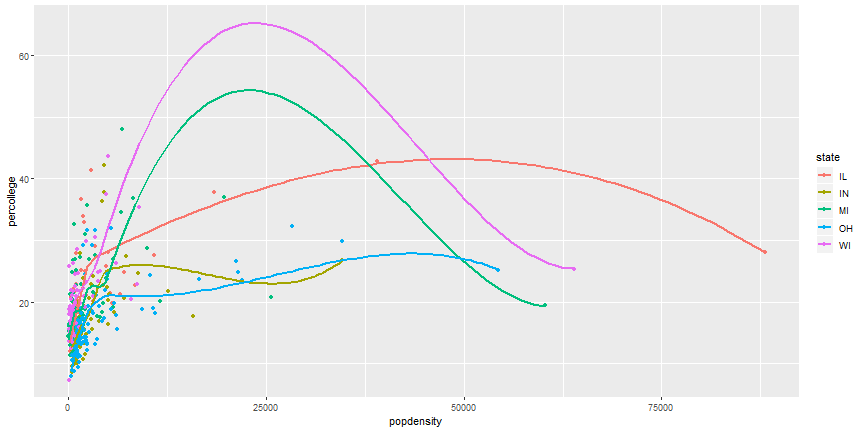
# Your Turn
1. It is possible to combine geoms, which we will do next, but try it first. Try to recreate this plot.
# Layered ggplot
```r
ggplot(midwest) +
geom_point(aes(x = popdensity, y = percollege, color = state)) +
geom_smooth(aes(x = popdensity, y = percollege, color = state),
se = FALSE)
```
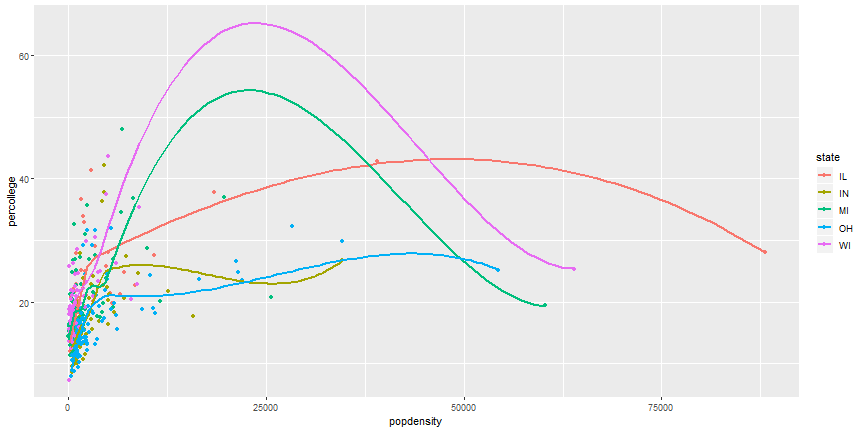
# Remove duplicate aesthetics
```r
ggplot(midwest,
aes(x = popdensity, y = percollege, color = state)) +
geom_point() +
geom_smooth(se = FALSE)
```
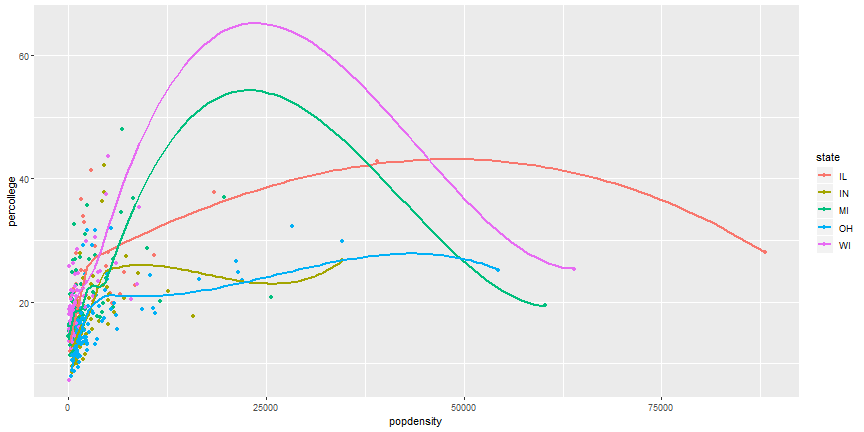
# Your Turn
1. Can you recreate the following figure?
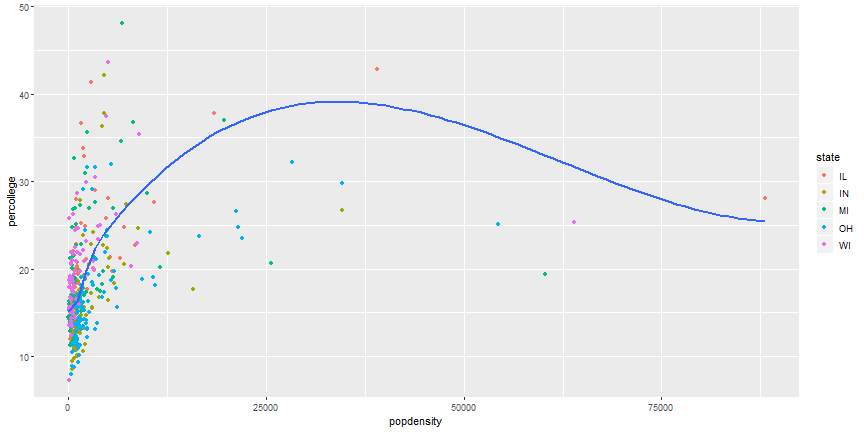
# Brief plot customization
```r
ggplot(midwest,
aes(x = popdensity, y = percollege, color = state)) +
geom_point() +
scale_x_continuous("Population Density",
breaks = seq(0, 80000, 20000)) +
scale_y_continuous("Percent College Graduates") +
scale_color_discrete("State")
```
# Brief plot customization Output

# Additional ggplot2 resources
+ ggplot2 website: <http://docs.ggplot2.org/current/index.html>
+ ggplot2 book: <http://www.springer.com/us/book/9780387981413>
+ R graphics cookbook: <http://www.cookbook-r.com/Graphs/>
# R works as a calculator
```r
1 + 2 - 3
```
```
## [1] 0
```
```r
5 * 7
```
```
## [1] 35
```
```r
2/1
```
```
## [1] 2
```
# R Calculator 2
```r
sqrt(4)
```
```
## [1] 2
```
```r
2^2
```
```
## [1] 4
```
# Can save objects to use later
```r
x <- 1 + 3
x
```
```
## [1] 4
```
```r
x * 3
```
```
## [1] 12
```
# R is case sensitive
```r
case_sensitive <- 10
Case_sensitive
```
```
## Error in eval(expr, envir, enclos): object 'Case_sensitive' not found
```
# R Functions
```r
set.seed(1)
rnorm(n = 5, mean = 0, sd = 1)
```
```
## [1] -0.6264538 0.1836433 -0.8356286 1.5952808 0.3295078
```
```r
set.seed(1)
rnorm(5, 0, 1)
```
```
## [1] -0.6264538 0.1836433 -0.8356286 1.5952808 0.3295078
```
```r
set.seed(1)
rnorm(sd = 1, n = 5, mean = 0)
```
```
## [1] -0.6264538 0.1836433 -0.8356286 1.5952808 0.3295078
```
# Working through Errors
1. Use `?function_name` to explore the details of the function. The examples at the bottom of every R help page can be especially helpful.
+ <https://www.rdocumentation.org/> provided by DataCamp is a great alternative as well.
2. If this does not help, copy and paste the error and search on the internet.
# Using `dplyr` for data manipulation
The `dplyr` package uses verbs for common data manipulation tasks. These include:
- `filter()`
- `count()`
- `arrange()`
- `select()`
- `mutate()`
- `summarise()`
# Data
<https://fivethirtyeight.com/features/both-republicans-and-democrats-have-an-age-problem/>
```r
install.packages('fivethirtyeight')
library(fivethirtyeight)
```
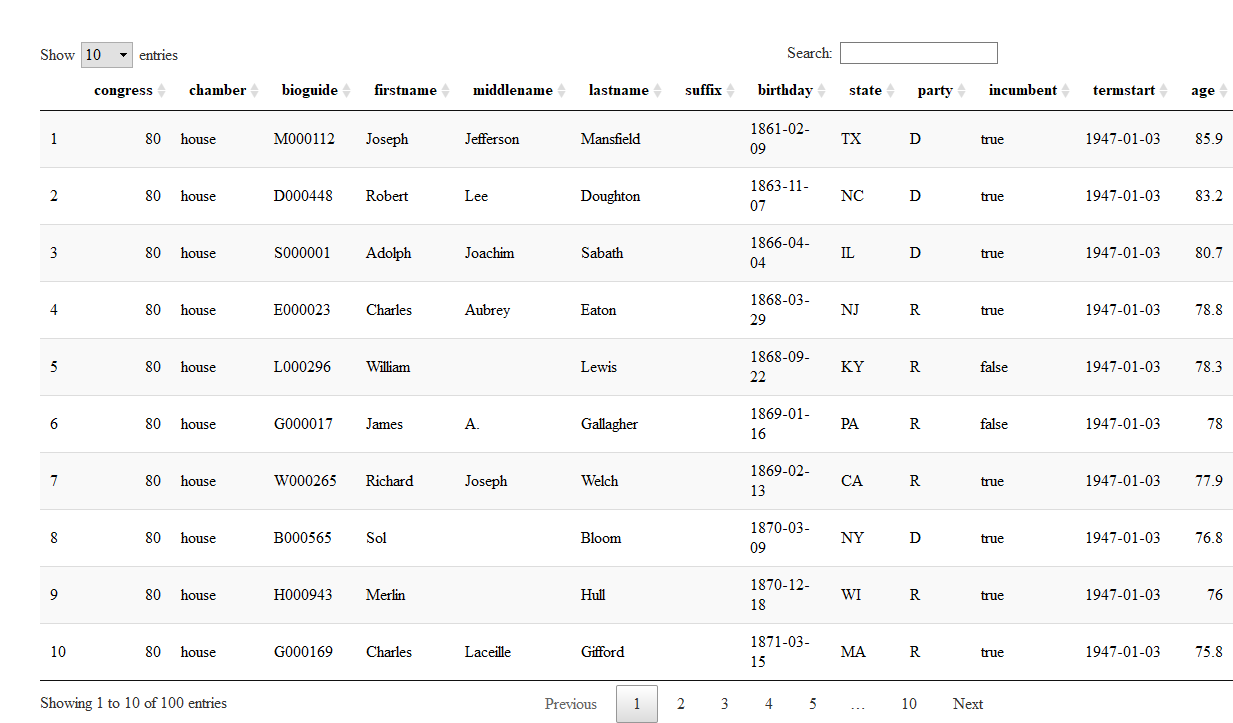
# Using `filter`
```r
filter(congress_age, congress == 80)
```
```
## # A tibble: 555 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 545 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Save filtered results to object
```r
congress_80 <- filter(congress_age, congress == 80)
```
# Other operators for numbers
- `>`
- `<`
- `>=`
- `<=`
# Your Turn
1. Select all rows where the congress member was older than 80 at the start of the term.
2. Use the `is.na` function to identify congress members that have missing middlenames.
# Filter character variables
```r
senate <- filter(congress_age, chamber == 'senate')
```
# Combine Operations - AND
```r
filter(congress_age, congress == 80, chamber == 'senate')
```
```
## # A tibble: 102 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 senate C000133 Arthur <NA> Capper <NA>
## 2 80 senate G000418 Theodore Francis Green <NA>
## 3 80 senate M000499 Kenneth Douglas McKellar <NA>
## 4 80 senate R000112 Clyde Martin Reed <NA>
## 5 80 senate M000895 Edward Hall Moore <NA>
## 6 80 senate O000146 John Holmes Overton <NA>
## 7 80 senate M001108 James Edward Murray <NA>
## 8 80 senate M000308 Patrick Anthony McCarran <NA>
## 9 80 senate T000165 Elmer <NA> Thomas <NA>
## 10 80 senate W000021 Robert Ferdinand Wagner <NA>
## # ... with 92 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Equivalent AND Statement
```r
filter(congress_age, congress == 80 & chamber == 'senate')
```
```
## # A tibble: 102 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 senate C000133 Arthur <NA> Capper <NA>
## 2 80 senate G000418 Theodore Francis Green <NA>
## 3 80 senate M000499 Kenneth Douglas McKellar <NA>
## 4 80 senate R000112 Clyde Martin Reed <NA>
## 5 80 senate M000895 Edward Hall Moore <NA>
## 6 80 senate O000146 John Holmes Overton <NA>
## 7 80 senate M001108 James Edward Murray <NA>
## 8 80 senate M000308 Patrick Anthony McCarran <NA>
## 9 80 senate T000165 Elmer <NA> Thomas <NA>
## 10 80 senate W000021 Robert Ferdinand Wagner <NA>
## # ... with 92 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Filter - OR
```r
filter(congress_age, congress == 80 | congress == 81)
```
```
## # A tibble: 1,112 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 1,102 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# `%in%`
```r
filter(congress_age, congress %in% c(80, 81))
```
```
## # A tibble: 1,112 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 1,102 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Not Operator
```r
filter(congress_age, congress != 80)
```
```
## # A tibble: 18,080 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 81 house D000448 Robert Lee Doughton <NA>
## 2 81 house S000001 Adolph Joachim Sabath <NA>
## 3 81 house E000023 Charles Aubrey Eaton <NA>
## 4 81 house W000265 Richard Joseph Welch <NA>
## 5 81 house B000565 Sol <NA> Bloom <NA>
## 6 81 house H000943 Merlin <NA> Hull <NA>
## 7 81 house B000545 Schuyler Otis Bland <NA>
## 8 81 house K000138 John Hosea Kerr <NA>
## 9 81 house C000932 Robert <NA> Crosser <NA>
## 10 81 house K000039 John <NA> Kee <NA>
## # ... with 18,070 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Not Operator 2
```r
filter(congress_age, congress == 80 & !chamber == 'senate')
```
```
## # A tibble: 453 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 443 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Working with logicals
```r
filter(congress_age, incumbent)
```
```
## # A tibble: 15,698 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house W000265 Richard Joseph Welch <NA>
## 6 80 house B000565 Sol <NA> Bloom <NA>
## 7 80 house H000943 Merlin <NA> Hull <NA>
## 8 80 house G000169 Charles Laceille Gifford <NA>
## 9 80 house B000545 Schuyler Otis Bland <NA>
## 10 80 house R000358 John Marshall Robsion <NA>
## # ... with 15,688 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Your Turn
1. Select the Senators from Iowa.
2. Select the Senators from Iowa that are not inbumbents.
# All boolean options
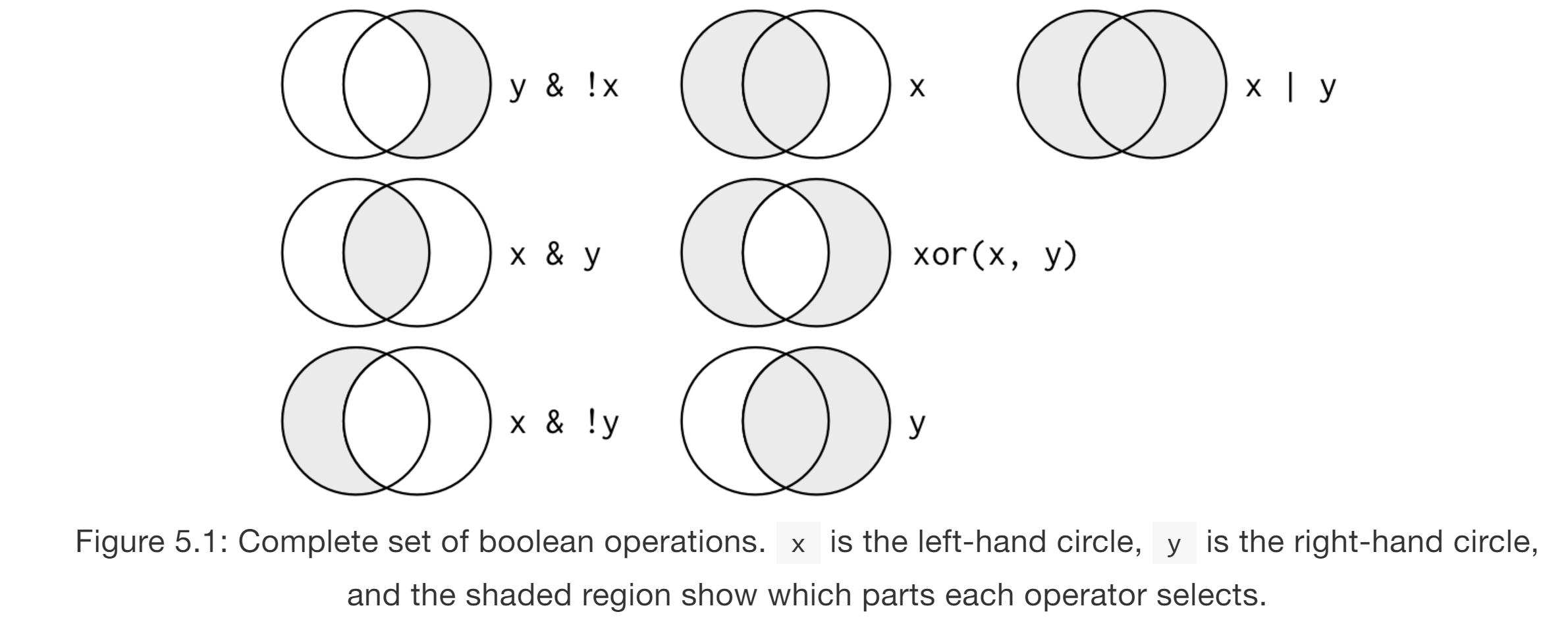
# Using `count`
```r
count(congress_age, party)
```
```
## # A tibble: 6 x 2
## party n
## <chr> <int>
## 1 AL 3
## 2 D 10290
## 3 I 63
## 4 ID 4
## 5 L 1
## 6 R 8274
```
```r
count(congress_age, incumbent)
```
```
## # A tibble: 2 x 2
## incumbent n
## <lgl> <int>
## 1 FALSE 2937
## 2 TRUE 15698
```
# Using `arrange`
```r
arrange(congress_age, state)
```
```
## # A tibble: 18,635 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house B000201 Edward Lewis Bartlett <NA>
## 2 81 house B000201 Edward Lewis Bartlett <NA>
## 3 82 house B000201 Edward Lewis Bartlett <NA>
## 4 83 house B000201 Edward Lewis Bartlett <NA>
## 5 84 house B000201 Edward Lewis Bartlett <NA>
## 6 85 house B000201 Edward Lewis Bartlett <NA>
## 7 86 house R000282 Ralph Julian Rivers <NA>
## 8 86 senate G000508 Ernest <NA> Gruening <NA>
## 9 86 senate B000201 Edward Lewis Bartlett <NA>
## 10 87 house R000282 Ralph Julian Rivers <NA>
## # ... with 18,625 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Add more variables
```r
arrange(congress_age, state, party)
```
```
## # A tibble: 18,635 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house B000201 Edward Lewis Bartlett <NA>
## 2 81 house B000201 Edward Lewis Bartlett <NA>
## 3 82 house B000201 Edward Lewis Bartlett <NA>
## 4 83 house B000201 Edward Lewis Bartlett <NA>
## 5 84 house B000201 Edward Lewis Bartlett <NA>
## 6 85 house B000201 Edward Lewis Bartlett <NA>
## 7 86 house R000282 Ralph Julian Rivers <NA>
## 8 86 senate G000508 Ernest <NA> Gruening <NA>
## 9 86 senate B000201 Edward Lewis Bartlett <NA>
## 10 87 house R000282 Ralph Julian Rivers <NA>
## # ... with 18,625 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Descending Order
```r
arrange(congress_age, desc(congress))
```
```
## # A tibble: 18,635 x 13
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 113 house H000067 Ralph M. Hall <NA>
## 2 113 house D000355 John D. Dingell <NA>
## 3 113 house C000714 John <NA> Conyers Jr.
## 4 113 house S000480 Louise McIntosh Slaughter <NA>
## 5 113 house R000053 Charles B. Rangel <NA>
## 6 113 house J000174 Sam Robert Johnson <NA>
## 7 113 house Y000031 C. W. Bill Young <NA>
## 8 113 house C000556 Howard <NA> Coble <NA>
## 9 113 house L000263 Sander M. Levin <NA>
## 10 113 house Y000033 Don E. Young <NA>
## # ... with 18,625 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Your Turn
1. Count the number of congress members from each party that are older than 80 at term start.
2. Arrange the result from above by the variable n.
# Using `select`
```r
select(congress_age, congress, chamber, party, age)
```
```
## # A tibble: 18,635 x 4
## congress chamber party age
## <int> <chr> <chr> <dbl>
## 1 80 house D 85.9
## 2 80 house D 83.2
## 3 80 house D 80.7
## 4 80 house R 78.8
## 5 80 house R 78.3
## 6 80 house R 78.0
## 7 80 house R 77.9
## 8 80 house D 76.8
## 9 80 house R 76.0
## 10 80 house R 75.8
## # ... with 18,625 more rows
```
# Helper functions
- `starts_with()`
- `ends_with()`
- `contains()`
- `matches()`
- `num_range()`
- `:`
- `everything()`
# `starts_with` helper
```r
select(congress_age, starts_with('s'))
```
```
## # A tibble: 18,635 x 2
## suffix state
## <chr> <chr>
## 1 <NA> TX
## 2 <NA> NC
## 3 <NA> IL
## 4 <NA> NJ
## 5 <NA> KY
## 6 <NA> PA
## 7 <NA> CA
## 8 <NA> NY
## 9 <NA> WI
## 10 <NA> MA
## # ... with 18,625 more rows
```
# Contains helper
```r
select(congress_age, contains('name'))
```
```
## # A tibble: 18,635 x 3
## firstname middlename lastname
## <chr> <chr> <chr>
## 1 Joseph Jefferson Mansfield
## 2 Robert Lee Doughton
## 3 Adolph Joachim Sabath
## 4 Charles Aubrey Eaton
## 5 William <NA> Lewis
## 6 James A. Gallagher
## 7 Richard Joseph Welch
## 8 Sol <NA> Bloom
## 9 Merlin <NA> Hull
## 10 Charles Laceille Gifford
## # ... with 18,625 more rows
```
# Colon
```r
select(congress_age, congress:birthday)
```
```
## # A tibble: 18,635 x 8
## congress chamber bioguide firstname middlename lastname suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 18,625 more rows, and 1 more variables: birthday <date>
```
# Drop variables
```r
select(congress_age, -firstname, -state, -party, -incumbent, -chamber)
```
```
## # A tibble: 18,635 x 8
## congress bioguide middlename lastname suffix birthday termstart
## <int> <chr> <chr> <chr> <chr> <date> <date>
## 1 80 M000112 Jefferson Mansfield <NA> 1861-02-09 1947-01-03
## 2 80 D000448 Lee Doughton <NA> 1863-11-07 1947-01-03
## 3 80 S000001 Joachim Sabath <NA> 1866-04-04 1947-01-03
## 4 80 E000023 Aubrey Eaton <NA> 1868-03-29 1947-01-03
## 5 80 L000296 <NA> Lewis <NA> 1868-09-22 1947-01-03
## 6 80 G000017 A. Gallagher <NA> 1869-01-16 1947-01-03
## 7 80 W000265 Joseph Welch <NA> 1869-02-13 1947-01-03
## 8 80 B000565 <NA> Bloom <NA> 1870-03-09 1947-01-03
## 9 80 H000943 <NA> Hull <NA> 1870-12-18 1947-01-03
## 10 80 G000169 Laceille Gifford <NA> 1871-03-15 1947-01-03
## # ... with 18,625 more rows, and 1 more variables: age <dbl>
```
# Reorder with `everything`
```r
select(congress_age, congress, chamber, incumbent, age, everything())
```
```
## # A tibble: 18,635 x 13
## congress chamber incumbent age bioguide firstname middlename
## <int> <chr> <lgl> <dbl> <chr> <chr> <chr>
## 1 80 house TRUE 85.9 M000112 Joseph Jefferson
## 2 80 house TRUE 83.2 D000448 Robert Lee
## 3 80 house TRUE 80.7 S000001 Adolph Joachim
## 4 80 house TRUE 78.8 E000023 Charles Aubrey
## 5 80 house FALSE 78.3 L000296 William <NA>
## 6 80 house FALSE 78.0 G000017 James A.
## 7 80 house TRUE 77.9 W000265 Richard Joseph
## 8 80 house TRUE 76.8 B000565 Sol <NA>
## 9 80 house TRUE 76.0 H000943 Merlin <NA>
## 10 80 house TRUE 75.8 G000169 Charles Laceille
## # ... with 18,625 more rows, and 6 more variables: lastname <chr>,
## # suffix <chr>, birthday <date>, state <chr>, party <chr>,
## # termstart <date>
```
# `rename` function
```r
rename(congress_age, first_name = firstname, last_name = lastname)
```
```
## # A tibble: 18,635 x 13
## congress chamber bioguide first_name middlename last_name suffix
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 18,625 more rows, and 6 more variables: birthday <date>,
## # state <chr>, party <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# Your Turn
1. Using the `dplyr` helper functions, select all the variables that start with the letter 'c'.
2. Rename the first three variables in the congress data to 'x1', 'x2', 'x3'.
3. After renaming the first three variables, use this new data (ensure you saved the previous step to an object) to select these three variables with the `num_range` function.
# Using `mutate`
```r
congress_red <- select(congress_age, congress, chamber, state, party)
mutate(congress_red,
democrat = ifelse(party == 'D', 1, 0),
num_democrat = sum(democrat)
)
```
```
## # A tibble: 18,635 x 6
## congress chamber state party democrat num_democrat
## <int> <chr> <chr> <chr> <dbl> <dbl>
## 1 80 house TX D 1 10290
## 2 80 house NC D 1 10290
## 3 80 house IL D 1 10290
## 4 80 house NJ R 0 10290
## 5 80 house KY R 0 10290
## 6 80 house PA R 0 10290
## 7 80 house CA R 0 10290
## 8 80 house NY D 1 10290
## 9 80 house WI R 0 10290
## 10 80 house MA R 0 10290
## # ... with 18,625 more rows
```
# Your Turn
1. Using the `diamonds` data, use `?diamonds` for more information on the data, use the `mutate` function to calculate the price per carat. Hint, this operation would involve standardizing the price variable so that all are comparable at 1 carat.
2. Using `mutate`, calculate the rank of the original price variable and the new price variable calculated above using the `min_rank` function. Are there differences in the ranking of the prices?
# Using `summarise`
```r
congress_2 <- mutate(congress_age,
democrat = ifelse(party == 'D', 1, 0)
)
summarise(congress_2,
num_democrat = sum(democrat)
)
```
```
## # A tibble: 1 x 1
## num_democrat
## <dbl>
## 1 10290
```
# `group_by`
```r
congress_grp <- group_by(congress_2, congress)
summarise(congress_grp,
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
```
```
## # A tibble: 34 x 4
## congress num_democrat total prop_democrat
## <int> <dbl> <int> <dbl>
## 1 80 247 555 0.4450450
## 2 81 330 557 0.5924596
## 3 82 292 555 0.5261261
## 4 83 274 557 0.4919210
## 5 84 288 544 0.5294118
## 6 85 295 547 0.5393053
## 7 86 356 554 0.6425993
## 8 87 339 559 0.6064401
## 9 88 332 552 0.6014493
## 10 89 371 548 0.6770073
## # ... with 24 more rows
```
# Explore trend
```r
num_dem <- summarise(congress_grp,
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
ggplot(num_dem, aes(x = congress, y = prop_democrat)) +
geom_line()
```
# Explore trend output
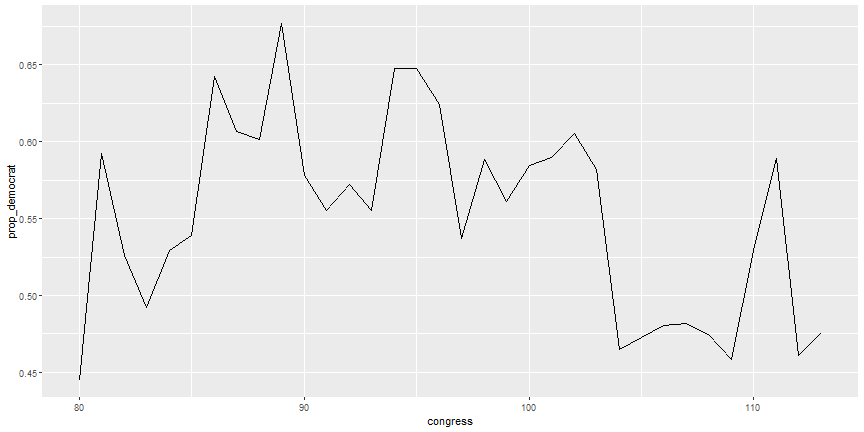
# Your Turn
1. Suppose we wanted to calculate the number and proportion of republicans instead of democrats, assuming these are the only two parties, edit the `summarise` command above to calculate these values.
2. Suppose instead of using `sum(democrat)` above, we used `mean(democrat)`, what does this value return? Why does it return this value?
# `group_by` with `mutate`
```r
congress_red <- select(congress_age, congress, chamber, state, party)
congress_grp <- group_by(congress_red, congress)
mutate(congress_grp,
democrat = ifelse(party == 'D', 1, 0),
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
```
# `group_by` with `mutate` output
```
## # A tibble: 18,635 x 8
## # Groups: congress [34]
## congress chamber state party democrat num_democrat total prop_democrat
## <int> <chr> <chr> <chr> <dbl> <dbl> <int> <dbl>
## 1 80 house TX D 1 247 555 0.445045
## 2 80 house NC D 1 247 555 0.445045
## 3 80 house IL D 1 247 555 0.445045
## 4 80 house NJ R 0 247 555 0.445045
## 5 80 house KY R 0 247 555 0.445045
## 6 80 house PA R 0 247 555 0.445045
## 7 80 house CA R 0 247 555 0.445045
## 8 80 house NY D 1 247 555 0.445045
## 9 80 house WI R 0 247 555 0.445045
## 10 80 house MA R 0 247 555 0.445045
## # ... with 18,625 more rows
```
# Chaining operations
```r
summarise(
group_by(
mutate(
filter(
congress_age, congress >= 100
),
democrat = ifelse(party == 'D', 1, 0)
),
congress, chamber
),
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
```
# The pipe `%>%` is the answer
```r
congress_age %>%
filter(congress >= 100) %>%
mutate(democrat = ifelse(party == 'D', 1, 0)) %>%
group_by(congress, chamber) %>%
summarise(
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
```
# The two are identical
```r
pipe_congress <- congress_age %>%
filter(congress >= 100) %>%
mutate(democrat = ifelse(party == 'D', 1, 0)) %>%
group_by(congress, chamber) %>%
summarise(
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
nested_congress <- summarise(
group_by(
mutate(
filter(
congress_age, congress >= 100
),
democrat = ifelse(party == 'D', 1, 0)
),
congress, chamber
),
num_democrat = sum(democrat),
total = n(),
prop_democrat = num_democrat / total
)
identical(pipe_congress, nested_congress)
```
```
## [1] TRUE
```
# Your Turn
1. Look at the following nested code and determine what is being done. Then translate this code to use the pipe operator.
```r
summarise(
group_by(
mutate(
filter(
diamonds,
color %in% c('D', 'E', 'F') & cut %in% c('Fair', 'Good', 'Very Good')
),
f_color = ifelse(color == 'F', 1, 0),
vg_cut = ifelse(cut == 'Very Good', 1, 0)
),
clarity
),
avg = mean(carat),
sd = sd(carat),
avg_p = mean(price),
num = n(),
summary_f_color = mean(f_color),
summary_vg_cut = mean(vg_cut)
)
```
# Other Useful dplyr functions
There are a set of functions that can greatly simplify data operations. These functions end with:
+ `*_if`
+ `*_each`
+ `*_all`
+ `*_at`
# Simple example
```r
rename_if(congress_age, is.character, toupper)
```
```
## # A tibble: 18,635 x 13
## congress CHAMBER BIOGUIDE FIRSTNAME MIDDLENAME LASTNAME SUFFIX
## <int> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 80 house M000112 Joseph Jefferson Mansfield <NA>
## 2 80 house D000448 Robert Lee Doughton <NA>
## 3 80 house S000001 Adolph Joachim Sabath <NA>
## 4 80 house E000023 Charles Aubrey Eaton <NA>
## 5 80 house L000296 William <NA> Lewis <NA>
## 6 80 house G000017 James A. Gallagher <NA>
## 7 80 house W000265 Richard Joseph Welch <NA>
## 8 80 house B000565 Sol <NA> Bloom <NA>
## 9 80 house H000943 Merlin <NA> Hull <NA>
## 10 80 house G000169 Charles Laceille Gifford <NA>
## # ... with 18,625 more rows, and 6 more variables: birthday <date>,
## # STATE <chr>, PARTY <chr>, incumbent <lgl>, termstart <date>, age <dbl>
```
# More realistic example
```r
starwars %>%
group_by(species, gender) %>%
summarise_if(is.numeric, mean, na.rm = TRUE)
```
```
## # A tibble: 43 x 5
## # Groups: species [?]
## species gender height mass birth_year
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 Aleena male 79 15.00000 NaN
## 2 Besalisk male 198 102.00000 NaN
## 3 Cerean male 198 82.00000 92.0
## 4 Chagrian male 196 NaN NaN
## 5 Clawdite female 168 55.00000 NaN
## 6 Droid none 200 140.00000 15.0
## 7 Droid <NA> 120 46.33333 72.5
## 8 Dug male 112 40.00000 NaN
## 9 Ewok male 88 20.00000 8.0
## 10 Geonosian male 183 80.00000 NaN
## # ... with 33 more rows
```
# Can call more than one function
```r
starwars %>%
group_by(species, gender) %>%
summarise_if(is.numeric, funs(mean, median), na.rm = TRUE) %>%
add_tally()
```
```
## # A tibble: 43 x 9
## # Groups: species [38]
## species gender height_mean mass_mean birth_year_mean height_median
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Aleena male 79 15.00000 NaN 79
## 2 Besalisk male 198 102.00000 NaN 198
## 3 Cerean male 198 82.00000 92.0 198
## 4 Chagrian male 196 NaN NaN 196
## 5 Clawdite female 168 55.00000 NaN 168
## 6 Droid none 200 140.00000 15.0 200
## 7 Droid <NA> 120 46.33333 72.5 97
## 8 Dug male 112 40.00000 NaN 112
## 9 Ewok male 88 20.00000 8.0 88
## 10 Geonosian male 183 80.00000 NaN 183
## # ... with 33 more rows, and 3 more variables: mass_median <dbl>,
## # birth_year_median <dbl>, n <int>
```
# Read in your own data
- We will use data posted to GitHub: <https://github.com/lebebr01/iowa_data_science/tree/master/data>
# Data Import
```r
ufo <- read_csv("https://raw.githubusercontent.com/lebebr01/iowa_data_science/master/data/ufo.csv")
```
```
## Parsed with column specification:
## cols(
## `Date / Time` = col_character(),
## City = col_character(),
## State = col_character(),
## Shape = col_character(),
## Duration = col_character(),
## Summary = col_character(),
## Posted = col_character()
## )
```
# Show Data
```r
ufo
```
```
## # A tibble: 8,031 x 7
## `Date / Time` City State Shape Duration
## <chr> <chr> <chr> <chr> <chr>
## 1 12/12/14 17:30 North Wales PA Triangle 5 minutes
## 2 12/12/14 12:40 Cartersville GA Unknown 3.6 minutes
## 3 12/12/14 06:30 Isle of Man (UK/England) <NA> Light 2 seconds
## 4 12/12/14 01:00 Miamisburg OH Changing <NA>
## 5 12/12/14 00:00 Spotsylvania VA Unknown 1 minute
## 6 12/11/14 23:25 Kenner LA Chevron ~1 minute
## 7 12/11/14 23:15 Eugene OR Disk 2 minutes
## 8 12/11/14 20:04 Phoenix AZ Chevron 3 minutes
## 9 12/11/14 20:00 Franklin NC Disk 5 minutes
## 10 12/11/14 18:30 Longview WA Cylinder 10 seconds
## # ... with 8,021 more rows, and 2 more variables: Summary <chr>,
## # Posted <chr>
```
# Manual column names
```r
ufo_man <- read_csv("https://raw.githubusercontent.com/lebebr01/iowa_data_science/master/data/ufo.csv",
skip = 1,
col_names = c('Date/Time', 'City', 'State',
'Shape', 'Duration', 'Summary',
'Posted'))
```
```
## Parsed with column specification:
## cols(
## `Date/Time` = col_character(),
## City = col_character(),
## State = col_character(),
## Shape = col_character(),
## Duration = col_character(),
## Summary = col_character(),
## Posted = col_character()
## )
```
# Manual column names output
```r
ufo_man
```
```
## # A tibble: 8,031 x 7
## `Date/Time` City State Shape Duration
## <chr> <chr> <chr> <chr> <chr>
## 1 12/12/14 17:30 North Wales PA Triangle 5 minutes
## 2 12/12/14 12:40 Cartersville GA Unknown 3.6 minutes
## 3 12/12/14 06:30 Isle of Man (UK/England) <NA> Light 2 seconds
## 4 12/12/14 01:00 Miamisburg OH Changing <NA>
## 5 12/12/14 00:00 Spotsylvania VA Unknown 1 minute
## 6 12/11/14 23:25 Kenner LA Chevron ~1 minute
## 7 12/11/14 23:15 Eugene OR Disk 2 minutes
## 8 12/11/14 20:04 Phoenix AZ Chevron 3 minutes
## 9 12/11/14 20:00 Franklin NC Disk 5 minutes
## 10 12/11/14 18:30 Longview WA Cylinder 10 seconds
## # ... with 8,021 more rows, and 2 more variables: Summary <chr>,
## # Posted <chr>
```
# Manual column types
```r
ufo_date <- read_csv("Data/ufo.csv",
col_types = list(
'Date / Time' = col_datetime(format = "%m/%d/%y %H:%M"),
City = col_character(),
State = col_character(),
Shape = col_character(),
Duration = col_character(),
Summary = col_character(),
Posted = col_character()
))
```
```
## Warning in rbind(names(probs), probs_f): number of columns of result is not
## a multiple of vector length (arg 1)
```
```
## Warning: 56 parsing failures.
## row # A tibble: 5 x 5 col row col expected actual file expected <int> <chr> <chr> <chr> <chr> actual 1 119 Date / Time date like %m/%d/%y %H:%M 12/1/14 'Data/ufo.csv' file 2 194 Date / Time date like %m/%d/%y %H:%M 11/27/14 'Data/ufo.csv' row 3 236 Date / Time date like %m/%d/%y %H:%M 11/24/14 'Data/ufo.csv' col 4 407 Date / Time date like %m/%d/%y %H:%M 11/15/14 'Data/ufo.csv' expected 5 665 Date / Time date like %m/%d/%y %H:%M 10/31/14 'Data/ufo.csv'
## ... ................. ... .................................................................... ........ .................................................................... ...... .................................................................... .... .................................................................... ... .................................................................... ... .................................................................... ........ ....................................................................
## See problems(...) for more details.
```
# Manual column types output
```r
ufo_date
```
```
## # A tibble: 8,031 x 7
## `Date / Time` City State Shape Duration
## <dttm> <chr> <chr> <chr> <chr>
## 1 2014-12-12 17:30:00 North Wales PA Triangle 5 minutes
## 2 2014-12-12 12:40:00 Cartersville GA Unknown 3.6 minutes
## 3 2014-12-12 06:30:00 Isle of Man (UK/England) <NA> Light 2 seconds
## 4 2014-12-12 01:00:00 Miamisburg OH Changing <NA>
## 5 2014-12-12 00:00:00 Spotsylvania VA Unknown 1 minute
## 6 2014-12-11 23:25:00 Kenner LA Chevron ~1 minute
## 7 2014-12-11 23:15:00 Eugene OR Disk 2 minutes
## 8 2014-12-11 20:04:00 Phoenix AZ Chevron 3 minutes
## 9 2014-12-11 20:00:00 Franklin NC Disk 5 minutes
## 10 2014-12-11 18:30:00 Longview WA Cylinder 10 seconds
## # ... with 8,021 more rows, and 2 more variables: Summary <chr>,
## # Posted <chr>
```
# Still problems
```r
problems(ufo_date)
```
```
## # A tibble: 56 x 5
## row col expected actual file
## <int> <chr> <chr> <chr> <chr>
## 1 119 Date / Time date like %m/%d/%y %H:%M 12/1/14 'Data/ufo.csv'
## 2 194 Date / Time date like %m/%d/%y %H:%M 11/27/14 'Data/ufo.csv'
## 3 236 Date / Time date like %m/%d/%y %H:%M 11/24/14 'Data/ufo.csv'
## 4 407 Date / Time date like %m/%d/%y %H:%M 11/15/14 'Data/ufo.csv'
## 5 665 Date / Time date like %m/%d/%y %H:%M 10/31/14 'Data/ufo.csv'
## 6 797 Date / Time date like %m/%d/%y %H:%M 10/25/14 'Data/ufo.csv'
## 7 946 Date / Time date like %m/%d/%y %H:%M 10/19/14 'Data/ufo.csv'
## 8 1081 Date / Time date like %m/%d/%y %H:%M 10/14/14 'Data/ufo.csv'
## 9 1122 Date / Time date like %m/%d/%y %H:%M 10/12/14 'Data/ufo.csv'
## 10 1123 Date / Time date like %m/%d/%y %H:%M 10/12/14 'Data/ufo.csv'
## # ... with 46 more rows
```
# Other text formats
- tsv - tab separated files - `read_tsv`
- fixed width files - `read_fwf`
- white space generally - `read_table`
- delimiter generally - `read_delim`
# Your Turn
1. There is a tsv file posted on icon called "lotr_clean.tsv". Download this and read this data file into R.
2. Instead of specifying the path, use the function `file.choose()`. For example, `read_tsv(file.choose())`.
+ What does this function do?
+ Would you recommend this to be used in a reproducible document?
# Excel Files
```r
install.packages('readxl')
```
# `read_excel`
```r
library(readxl)
read_excel('data/titanic.xlsx')
```
```
## # A tibble: 891 x 12
## PassengerId Survived Pclass
## <dbl> <dbl> <dbl>
## 1 1 0 3
## 2 2 1 1
## 3 3 1 3
## 4 4 1 1
## 5 5 0 3
## 6 6 0 3
## 7 7 0 1
## 8 8 0 3
## 9 9 1 3
## 10 10 1 2
## # ... with 881 more rows, and 9 more variables: Name <chr>, Sex <chr>,
## # Age <dbl>, SibSp <dbl>, Parch <dbl>, Ticket <chr>, Fare <dbl>,
## # Cabin <chr>, Embarked <chr>
```
# Write Output Files
```r
ufo_count <- ufo %>%
group_by(State) %>%
mutate(num_state = n())
write_csv(ufo_count, path = 'path/to/save/file.csv')
```
# Additional Resources
+ R for Data Science: <http://r4ds.had.co.nz/>